Synoptic Intelligence Outperforms 20x Larger Language Model in Multi-Lable EHR Interpretation
Pairwise Meta-learning shows promise in preclinical, bioassay-centered molecular activity prediction. On top of this framework, we introduce a new element — synoptic learning — and build an advanced, synoptic intelligence engine for application in clinical studies and real-world healthcare.
A preliminary study is carried out on ICD disease labeling of de-identified electronic health records (EHR) and the performance of our synoptic intelligence is compared with Google’s T5 language model.
We will briefly introduce the rationale of study design before looking at the performance metrics:
1. Datasets
Foundation models, including T5, are known for a broad spectrum of applications by virtue of their ability to quickly learn from the limited data of each application. This advantage is attributed to the fact that the models have been pre-trained on a “large” dataset. In such a dataset, a high degree of content diversity is assumed.
T5 models were pre-trained on the C4 (Colossal Clean Crawled Corpus) dataset. It contains 365 million documents, summing to 156 billion tokens.
A subset of Medical Information Mart for Intensive Care III (MIMIC3) was used in a previous report to test different modules in Google’s T5 language models. We adopt the same data split in the current study.
The MIMIC3 data split yields 30k medical records for training, 10k for validation, and 10k for testing. This is about 12,167 times smaller than C4 in terms of per-record/per-document application.
2. Multi-label disease coding
Each MIMIC record is annotated with the medical conditions it involved in the form of ICD codes. The models are tested with their abilities to predict the presence/absence of 19 codes, which comprise the ICD9 disease & injury index, in each record’s annotation.
Notably, the presence of one index does not exclude the presence of another.
This reflects the reality in our daily healthcare where multiple medical conditions, such as co-morbidities and patients’ adverse reactions to the treatment, usually occur in the same case.
3. Model training on MIMIC3
As the baseline, the retrained T5-base encoder is fine-tuned. It begins overfitting after 10 epochs of fine-tuning on the relatively small, 30k training set.
The model of pairwise meta-learning is trained on the same 30k dataset from scratch.
For each model, the set of model parameters that yields the optimal performance on the validation set during the training process is used in testing.
4. Multi-label predictions from individual EHR sections
Each MIMIC record contains pre-defined sections.
11 relevant sections — Chief complaint, brief hospital course, history of present illness, past medical history, family history, social history, surgical history, medications on admission, physical exams, discharge medications, and discharge diagnosis — are used as inputs for the models to predict ICD labels.
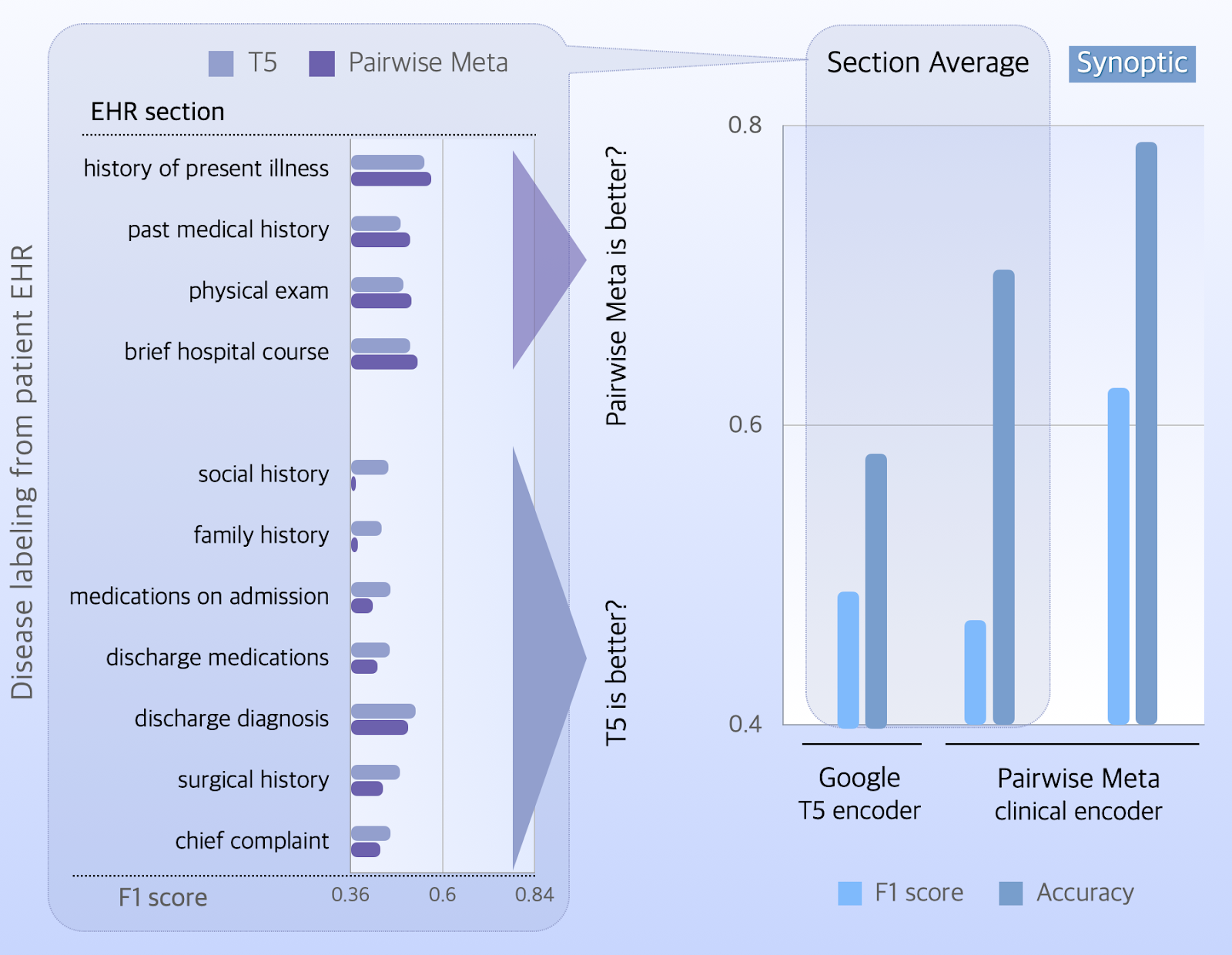
Just as in hospitals where medical encounters do not equally distribute across departments, data availability is not balanced over the 19 ICD codes. To attribute equal importance to all disease/injury categories, we adopt the macro average of F1 scores to present the model performance.
The T5 encoder has better macro-F1 scores on 7 EHR sections, and a section average of 0.49.
The model of pairwise meta-learning performs better on sections of medical history, hospital course, physical exams, and has a section-averaged score of 0.47.
In contrast to the marginally lower score of macro F1, the model of pairwise meta-learning has a better accuracy (0.7) than the T5 encoder (0.58), though.
Here comes the question:
Should we weigh the evidence on certain EHR sections or trust the average? Which scoring metric to consider?
Synoptic Intelligence Resolves Discrepancy in Clinical Statistics
When we enable predictions through the synoptic learning module, it captures hidden synergies of accessible EHR sections and improves both the scores of accuracy (0.79) and macro F1 (0.63) by an evident margin.
Contradicting statistics from different clinical studies and hospital encounters are a common issue in healthcare research and practice — not only as a consequence of sampling bias, type I/II errors, and missing data. The variability in real-world patient populations is also a major contributing factor.
Unlike drafting a scientific study report, we can’t conclude the issue like “this is a complex issue with multiple reasons; addressing this challenge requires a multi-faceted approach that includes A, B & C.” and leave the problem to the future.
In medical practice, we have to make decisions on the spot. The synoptic intelligence engine is developed to support real-time decision making.
Join Us to Solve The Problem in Real Life
It is noteworthy that synoptic intelligence improves model performance by 36%, as compared to the T5 encoder, using a 20x smaller parameter set (66MB vs 1.3GB). Striking escalation of the predicted power is anticipated when data availability matches the model size at scale.
See our offers to leverage this leading-edge technology in therapeutic development and clinical decision, or let us know what you need.